Note
Go to the end to download the full example code.
Soot precursor formation with time-varying mass flow rate#
Simulation of fuel injection into a vitiated air mixture to show formation of soot precursors.
Demonstrates the use of a user-supplied function for the mass flow rate through
a MassFlowController, and the use of the SolutionArray class to store results
during reactor network integration and use these results to generate plots.
Requires: cantera >= 3.2, matplotlib >= 2.0
import numpy as np
import matplotlib.pyplot as plt
import cantera as ct
# Use a reduced n-dodecane mechanism with PAH formation pathways
gas = ct.Solution('nDodecane_Reitz.yaml', 'nDodecane_IG')
gas.case_sensitive_species_names = True
# Create a Reservoir for the fuel inlet, set to pure dodecane
gas.TPX = 300, 20*ct.one_atm, 'c12h26:1.0'
inlet = ct.Reservoir(gas)
# Create Reactor and set initial contents to be products of lean combustion
gas.TP = 1000, 20*ct.one_atm
gas.set_equivalence_ratio(0.30, 'c12h26', 'n2:3.76, o2:1.0')
gas.equilibrate('TP')
r = ct.IdealGasReactor(gas)
r.volume = 0.001 # 1 liter
def fuel_mdot(total_mass=3.0e-3, std_dev=0.5, center_time=2.0):
"""Create a Gaussian pulse function."""
# units are kg for mass and seconds for times
amplitude = total_mass / (std_dev * np.sqrt(2 * np.pi))
fwhm = std_dev * 2 * np.sqrt(2 * np.log(2))
return ct.Func1("Gaussian", [amplitude, center_time, fwhm])
# Create an inlet for the fuel, supplied as a Gaussian pulse
mfc = ct.MassFlowController(inlet, r, mdot=fuel_mdot())
# Create the reactor network
sim = ct.ReactorNet([r])
# Integrate for 10 seconds, storing the results for later plotting
tfinal = 10.0
tnow = 0.0
i = 0
tprev = tnow
states = ct.SolutionArray(gas, extra=['t'])
while tnow < tfinal:
tnow = sim.step()
i += 1
# Storing results after every step can be excessive. Instead, store results
# every 10 steps, or more frequently if large steps are being taken.
if tnow-tprev > 1e-2 or i == 10:
i = 0
tprev = tnow
states.append(r.phase.state, t=tnow)
# nice names for species, including PAH species that can be considered
# as precursors to soot formation
species_aliases = {
'o2': 'O$_2$',
'h2o': 'H$_2$O',
'co': 'CO',
'co2': 'CO$_2$',
'h2': 'H$_2$',
'ch4': 'CH$_4$'
}
for name, alias in species_aliases.items():
gas.add_species_alias(name, alias)
pah_aliases = {
'A1c2h': 'phenylacetylene',
'A1c2h3': 'styrene',
'A1': 'benzene',
'A2': 'naphthalene',
'A2r5': 'acenaphthylene',
'A3': 'phenanthrene',
'A4': 'pyrene'
}
for name, alias in pah_aliases.items():
gas.add_species_alias(name, alias)
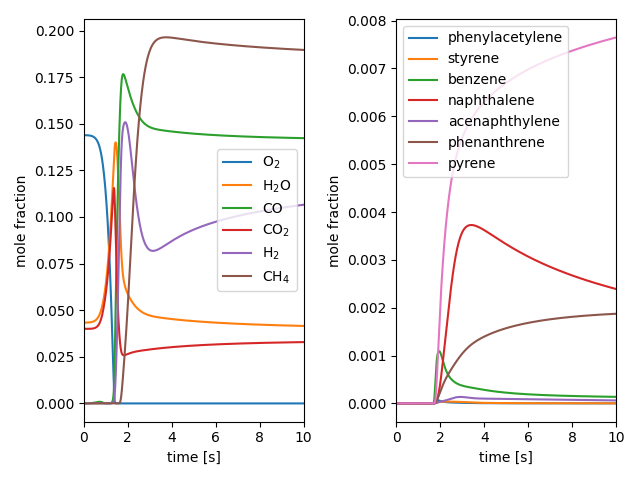
# Plot the concentrations of species of interest
f, ax = plt.subplots(1, 2)
for s in species_aliases.values():
ax[0].plot(states.t, states(s).X, label=s)
for s in pah_aliases.values():
ax[1].plot(states.t, states(s).X, label=s)
for a in ax:
a.legend(loc='best')
a.set_xlabel('time [s]')
a.set_ylabel('mole fraction')
a.set_xlim([0, tfinal])
f.tight_layout()
plt.show()
Total running time of the script: (0 minutes 0.983 seconds)

