Note
Go to the end to download the full example code.
Extracting a sub-mechanism#
An example demonstrating how to use Species and Reaction objects to programmatically extract a reaction submechanism. In this example, the CO/H2 oxidation reactions are extracted from the GRI 3.0 mechanism.
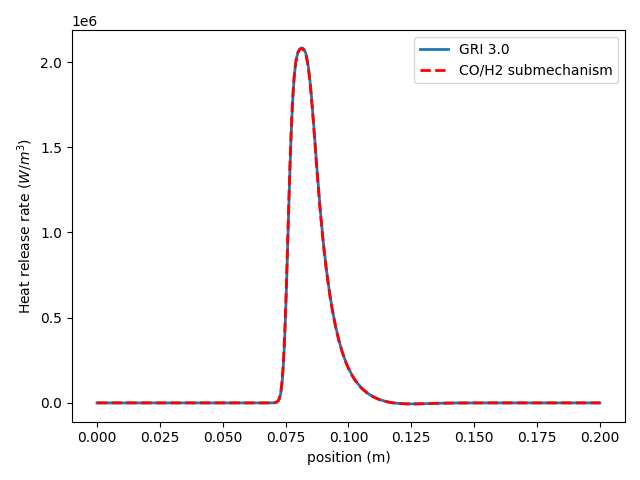
To test the submechanism, a premixed CO/H2 flame is simulated using the original mechanism and the submechanism, which demonstrates that the submechanism contains all of the important species and reactions.
Requires: cantera >= 2.6.0, matplotlib >= 2.0
from timeit import default_timer
import cantera as ct
import matplotlib.pyplot as plt
input_file = 'gri30.yaml'
all_species = ct.Species.list_from_file(input_file)
species = []
Filter species.
for S in all_species:
comp = S.composition
if 'C' in comp and 'H' in comp:
# Exclude all hydrocarbon species
continue
if 'N' in comp and comp != {'N': 2}:
# Exclude all nitrogen compounds except for N2
continue
if 'Ar' in comp:
# Exclude Argon
continue
species.append(S)
species_names = {S.name for S in species}
print('Species: {0}'.format(', '.join(S.name for S in species)))
Species: H2, H, O, O2, OH, H2O, HO2, H2O2, C, CO, CO2, N2
Filter reactions, keeping only those that only involve the selected species.
ref_phase = ct.Solution(thermo='ideal-gas', kinetics='gas', species=all_species)
all_reactions = ct.Reaction.list_from_file(input_file, ref_phase)
reactions = []
print('\nReactions:')
for R in all_reactions:
if not all(reactant in species_names for reactant in R.reactants):
continue
if not all(product in species_names for product in R.products):
continue
reactions.append(R)
print(R.equation)
print('\n')
gas1 = ct.Solution(input_file)
gas2 = ct.Solution(name="gri30-CO-H2-submech",
thermo="ideal-gas", kinetics="gas",
transport_model="mixture-averaged",
species=species, reactions=reactions)
Reactions:
2 O + M <=> O2 + M
H + O + M <=> OH + M
H2 + O <=> H + OH
HO2 + O <=> O2 + OH
H2O2 + O <=> HO2 + OH
CO + O (+M) <=> CO2 (+M)
CO + O2 <=> CO2 + O
H + O2 + M <=> HO2 + M
H + O2 + O2 <=> HO2 + O2
H + O2 + H2O <=> HO2 + H2O
H + O2 + N2 <=> HO2 + N2
H + O2 + AR <=> HO2 + AR
H + O2 <=> O + OH
2 H + M <=> H2 + M
2 H + H2 <=> H2 + H2
2 H + H2O <=> H2 + H2O
2 H + CO2 <=> H2 + CO2
H + OH + M <=> H2O + M
H + HO2 <=> H2O + O
H + HO2 <=> H2 + O2
H + HO2 <=> 2 OH
H + H2O2 <=> H2 + HO2
H + H2O2 <=> H2O + OH
H2 + OH <=> H + H2O
2 OH (+M) <=> H2O2 (+M)
2 OH <=> H2O + O
HO2 + OH <=> H2O + O2
H2O2 + OH <=> H2O + HO2
H2O2 + OH <=> H2O + HO2
C + OH <=> CO + H
CO + OH <=> CO2 + H
2 HO2 <=> H2O2 + O2
2 HO2 <=> H2O2 + O2
CO + HO2 <=> CO2 + OH
C + O2 <=> CO + O
HO2 + OH <=> H2O + O2
Save the resulting mechanism for later use.
gas2.update_user_header({"description": "CO-H2 submechanism extracted from GRI-Mech 3.0"})
gas2.write_yaml("gri30-CO-H2-submech.yaml", header=True)
Simulate a flame with each mechanism.
def solve_flame(gas):
gas.TPX = 373, 0.05*ct.one_atm, 'H2:0.4, CO:0.6, O2:1, N2:3.76'
# Create the flame simulation object
sim = ct.CounterflowPremixedFlame(gas=gas, width=0.2)
sim.reactants.mdot = 0.12 # kg/m^2/s
sim.products.mdot = 0.06 # kg/m^2/s
sim.set_refine_criteria(ratio=3, slope=0.1, curve=0.2)
sim.solve(0, auto=True)
return sim
t1 = default_timer()
sim1 = solve_flame(gas1)
t2 = default_timer()
print('Solved with GRI 3.0 in {0:.2f} seconds'.format(t2-t1))
sim2 = solve_flame(gas2)
t3 = default_timer()
print('Solved with CO/H2 submechanism in {0:.2f} seconds'.format(t3-t2))
Solved with GRI 3.0 in 7.34 seconds
Solved with CO/H2 submechanism in 0.37 seconds
Plot the results to show that the submechanism gives the same results as the original.
plt.plot(sim1.grid, sim1.heat_release_rate,
lw=2, label='GRI 3.0')
plt.plot(sim2.grid, sim2.heat_release_rate,
'r--', lw=2, label='CO/H2 submechanism')
plt.ylabel('Heat release rate ($W/m^3$)')
plt.xlabel('position (m)')
plt.legend(loc='upper right')
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 8.003 seconds)

